Sub-Project1: Structure determination for protein complexes using cryo-electron microscopy
-
-
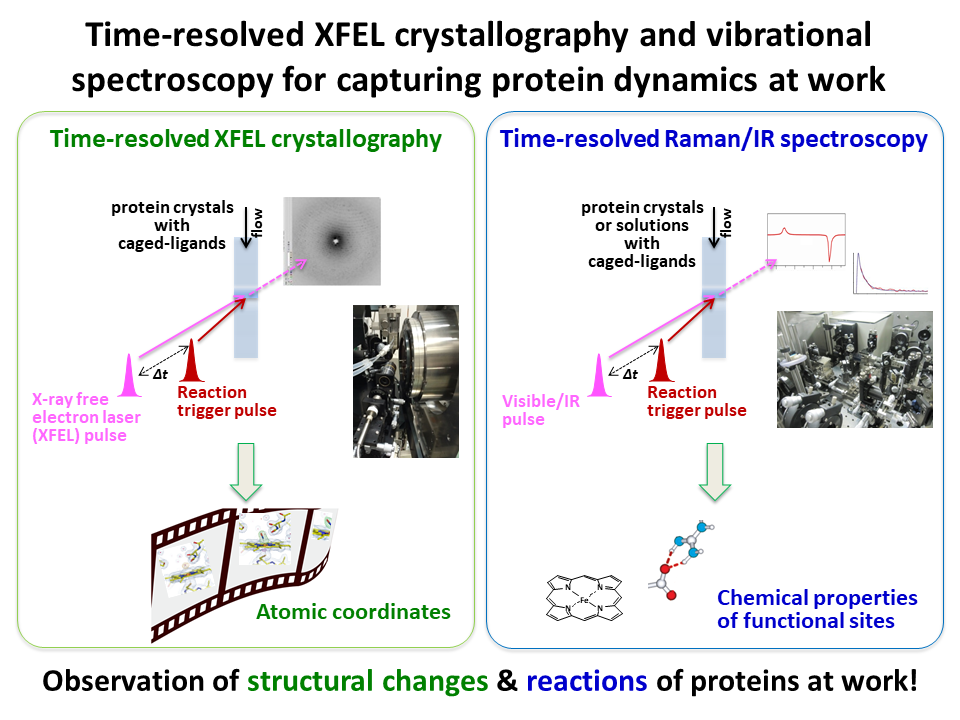
1.Time-resolved XFEL crystallography and vibrational spectroscopy for capturing protein dynamics at work
RIKEN SPring-8 Center, Imaging development Team
Senior Research Scientist: Minoru Kubo
We develop the time-resolved techniques of X-ray crystallography and vibrational spectroscopy, and study protein reaction dynamics at the atomic and electronic levels in real time.
-
-
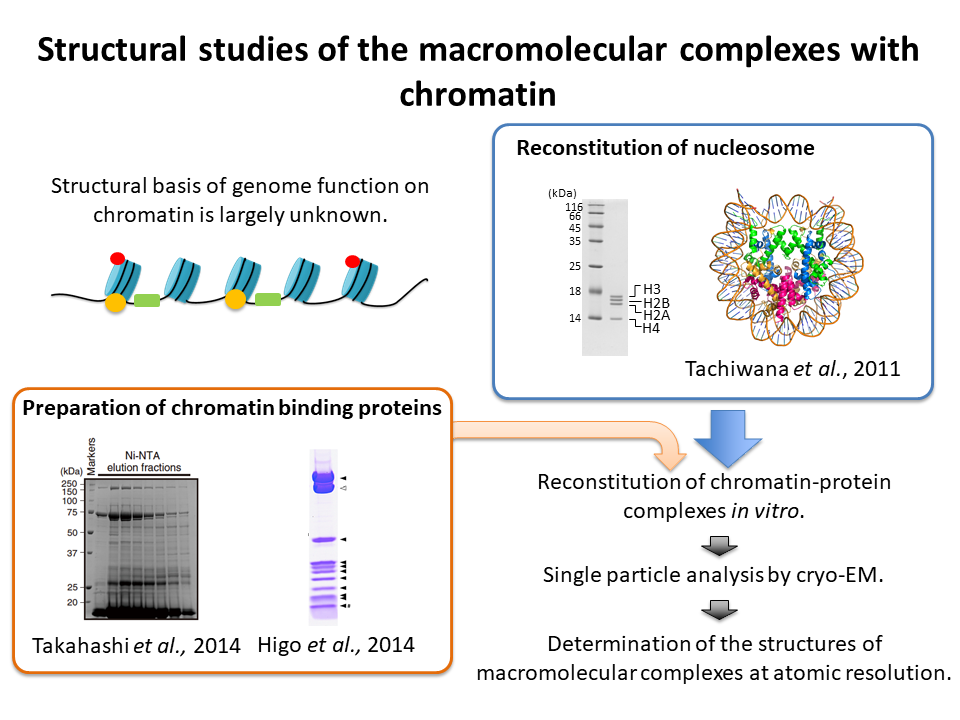
2.Structural studies of the macromolecular complexes with chromatin
Laboratory of structural biology
Team leader: Hitoshi Kurumizaka
We reconstitute the macromolecular complexes with chromatin and determine their three dimensional structures by cryo-EM.
-
-
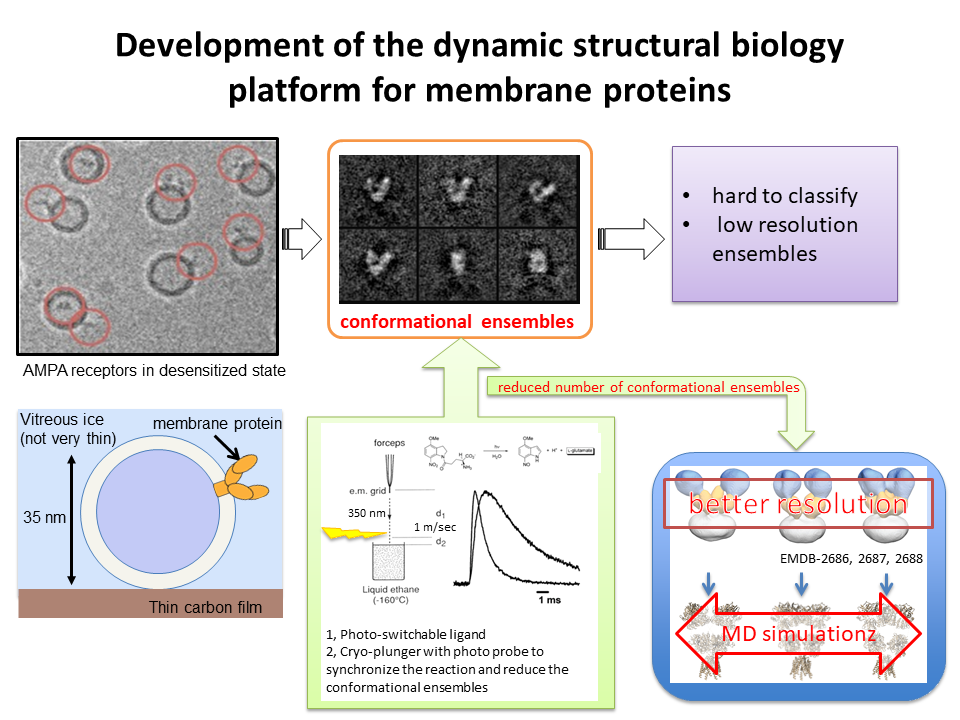
3.Development of the dynamic structural biology platform for membrane proteins
Center for Life Science Technologies, Protein Functional and Structural Biology Team
Senior Scientist: Hideki Shigematsu
We develop the new method in cryoEM single particle analysis to visualize conformational ensembles of membrane proteins in function.
-
-
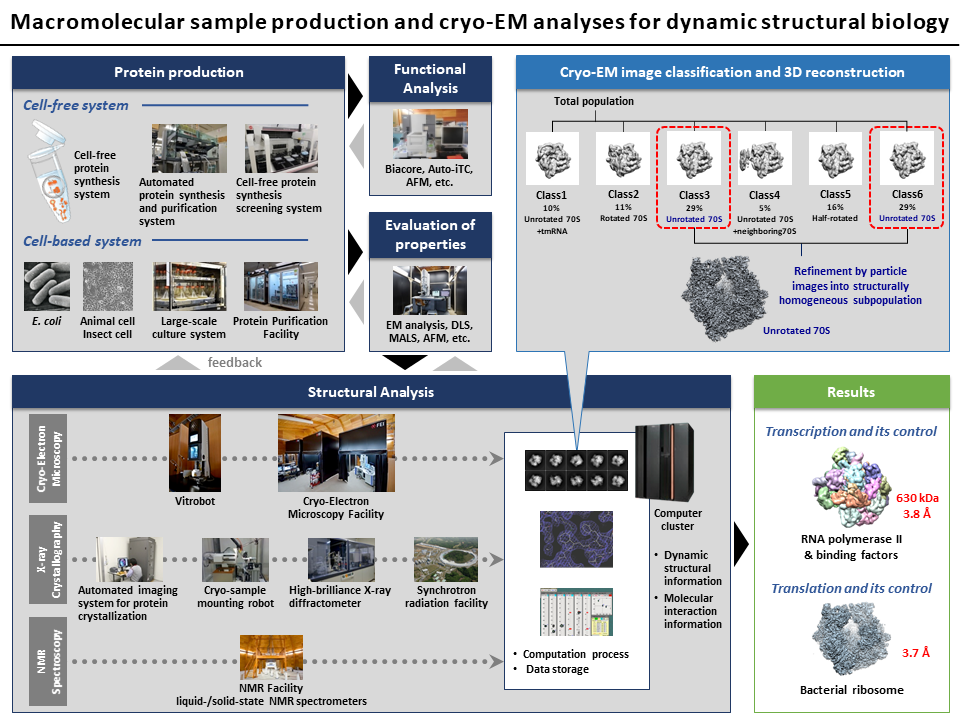
4.Macromolecular sample production and cryo-EM analyses for dynamic structural biology
RIKEN BDR Laboratory for Protein Functional and Structural Biology
Team leader: Mikako Shirouzu
We develop the methods for preparation of biomolecules reflecting their dynamic functional states and operate an integrated structural analysis pipeline including cryo-electron microscopy.
-
-
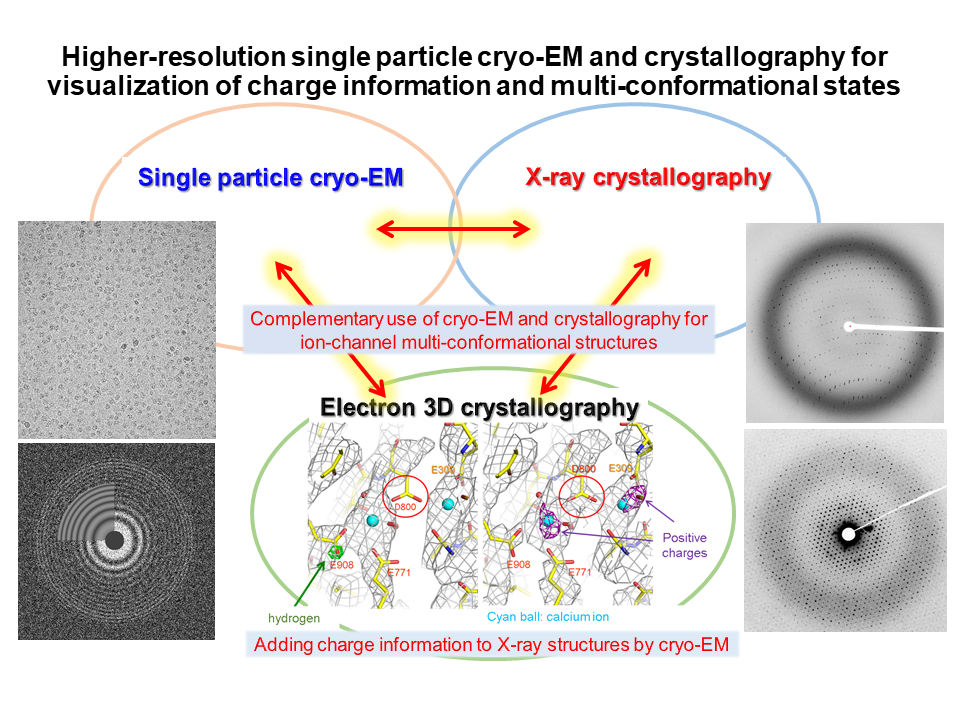
5.High-resolution single particle cryo-EM and crystallography of membrane protein complexes
Biostructural mechanism laboratory
Chief scientist: Koji Yonekura
We carry out single particle cryo-EM and crystallography of membrane protein complexes, and develop methodologies for higher-resolution cryo-EM and analyses of charge information and multi conformational states.
-
-
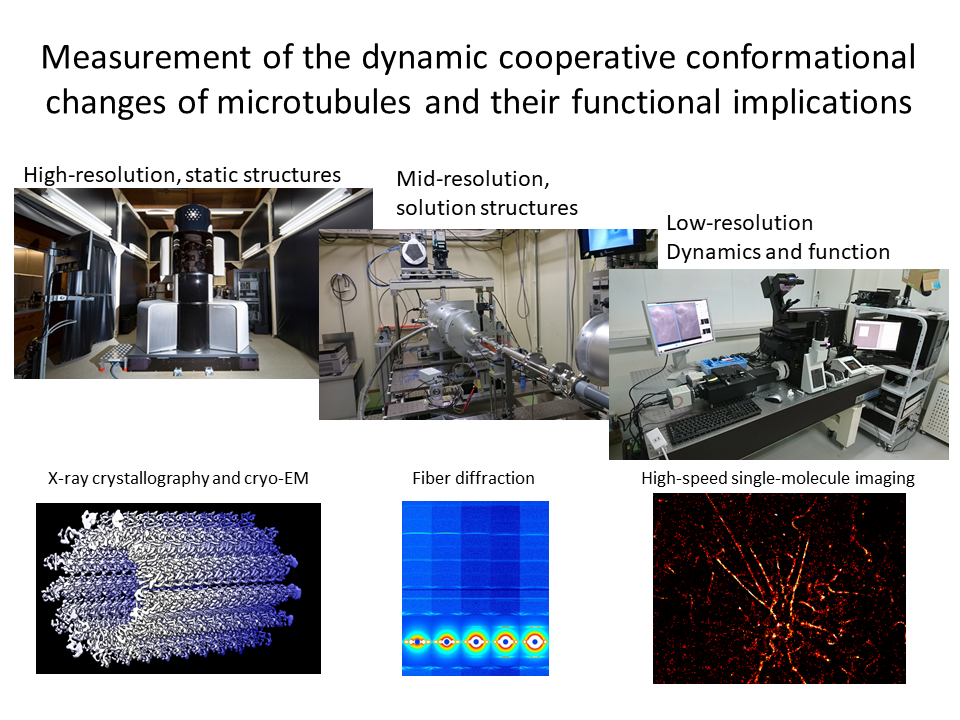
6.Cooperative conformational changes of microtubules and their functional implications
Laboratory for Cell Polarity Regulation
Team leader: Yasushi Okada
Microtubule is a dynamic cytoskeleton and playing various essential roles in living cells. To investigate their regulation, we examine the dynamic conformational changes of tubulin subunits in microtubules by using various measurement technologies.
Sub-Project2: Development of high-sensitivity NMR spectoroscopy
-
-
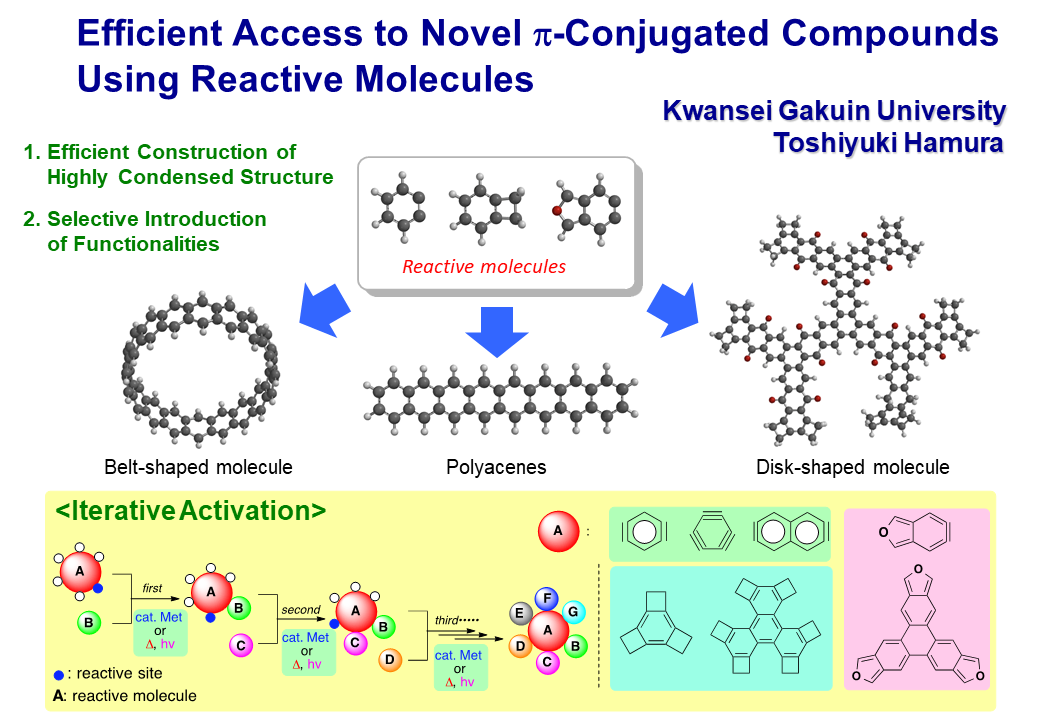
1.Efficient Synthetic Route to Novel Pi-Conjugated Molecules and Development of their Function,
Hamura Lab in Kwansei Gakuin University
Team leader: Toshiyuki Hamura
The purpose of this research is development of an efficient synthetic route to various pi-conjugated systems based on the reaction integration using the reactive molecules, having potentially attractive reactivities. Three kinds of activations by heat, light, and transition metal catalyst enable the selective construction of complex structures by introducing diverse functionalities.
-
-
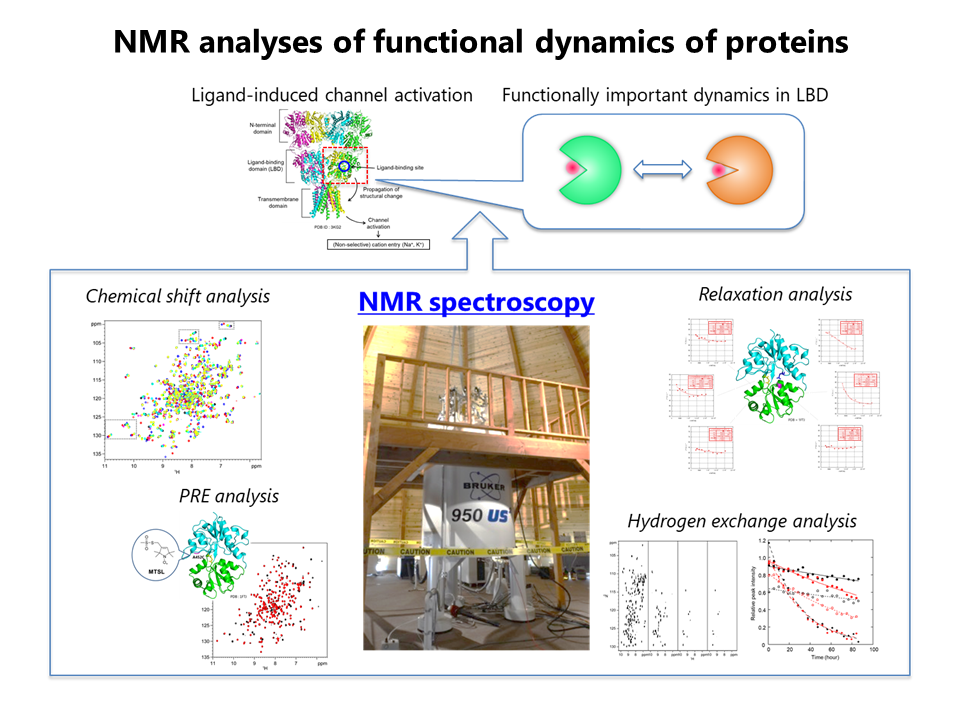
2.NMR analyses of functional dynamics of proteins / Triplet DNP application to membrane-related proteins.
Functional Structure Biology Laboratory, Graduate School of Medical Life Science, Yokohama City University
Team leader: Hideo Takahashi
We analyze functionally important but hidden dynamics of drug-target proteins by using NMR spectroscopy. We also tackle the triplet DNP application to membrane-related proteins.
-
-
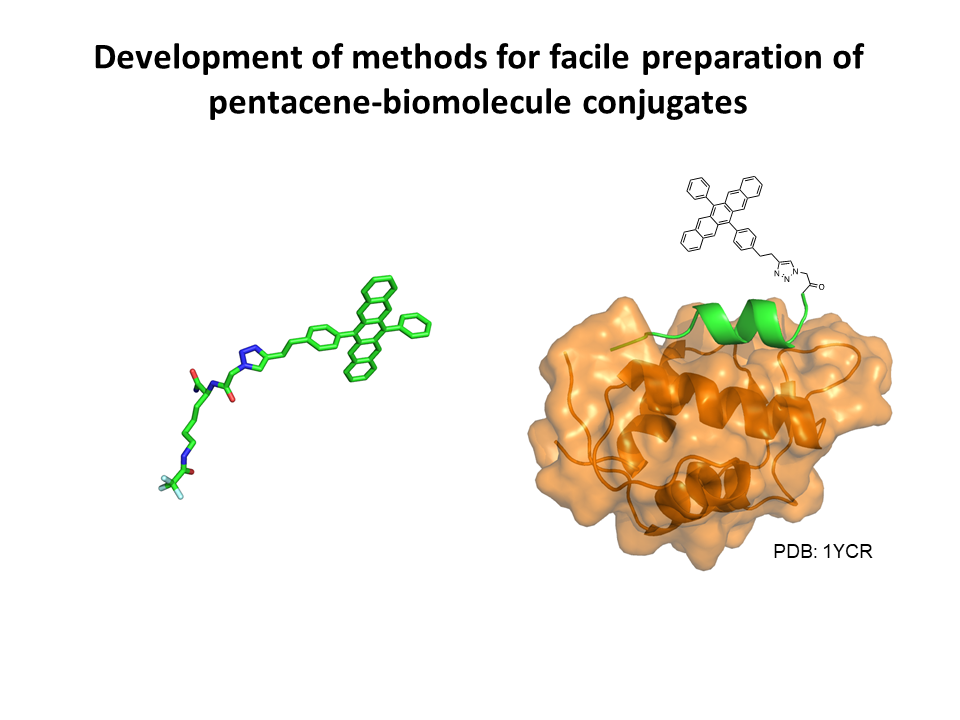
3.Development of methods for facile preparation of pentacene-biomolecule conjugates
Graduate School of Engineering, The University of Tokyo
Team leader: Shinsuke Sando
We have taken a challenge to develop a facile method to conjugate key molecule “Pentacene” with various biomolecules
-
-
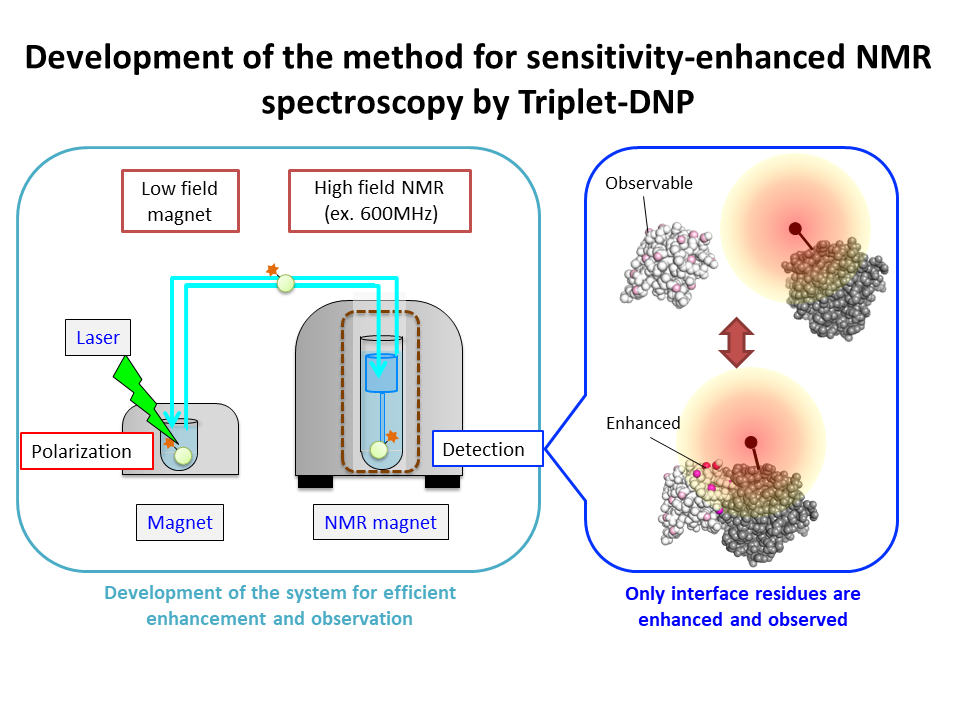
4.Development of the method for sensitivity-enhanced NMR spectroscopy by Triplet-DNP
Laboratory for Biomolecular Structure and Dynamics
Team leader: Takanori Kigawa
We will develop the new method for observing molecular interaction using sensitivity-enhanced NMR spectroscopy by Triplet-DNP.
-
-
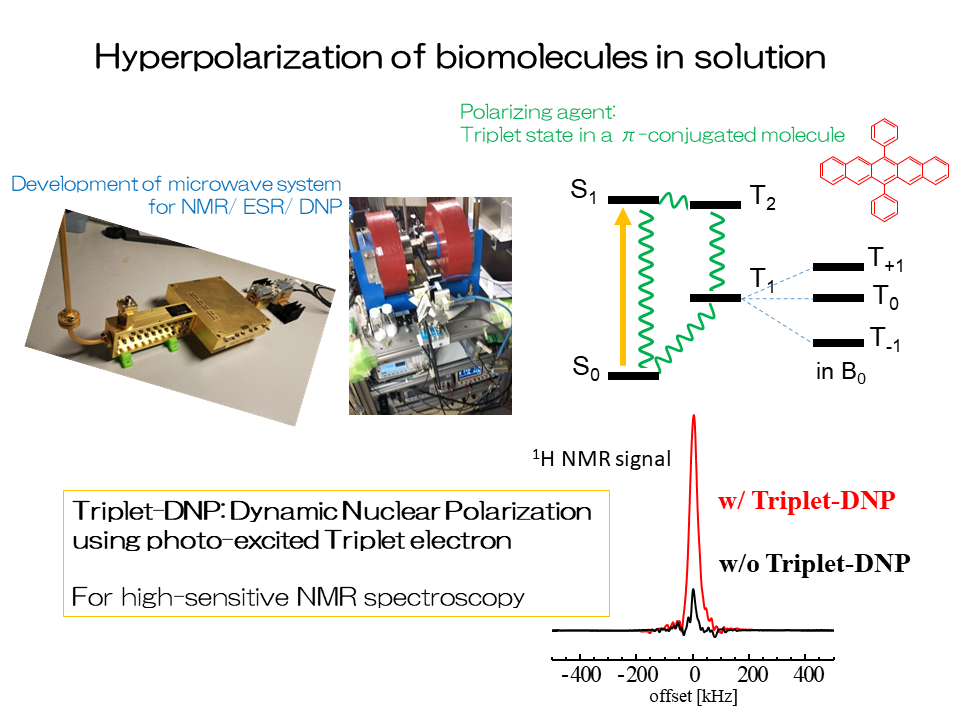
5.Hyperpolarization of biomolecules in solution
Uesaka Spin-isospin Laboratory
Team leader: Tomohiro Uesaka
We will develop a method to hyper-polarize biomolecules using laser and microwave for sensitivity-enhanced NMR spectroscopy.
Sub-Project3: Development of experimental-data-driven MD simulations
-
-
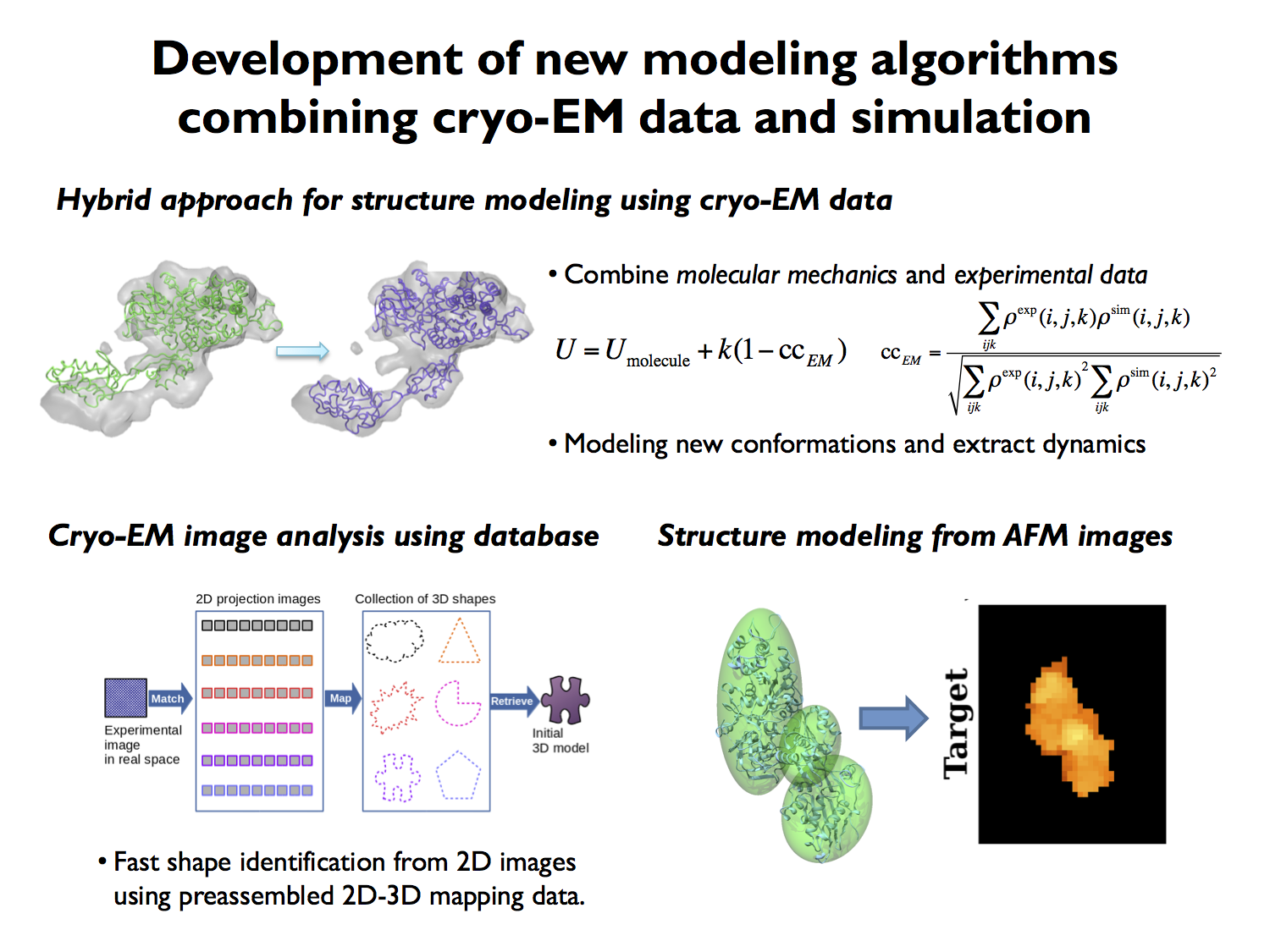
1.Development of new modeling algorithms combining cryo-EM data and simulation
R-CCS Computational Structural Biology Research Unit
Team leader: Florence Tama
We develop new algorithms to construct 3D models and obtain dynamical information from Cryo-EM experimental data of biological molecules
-
-
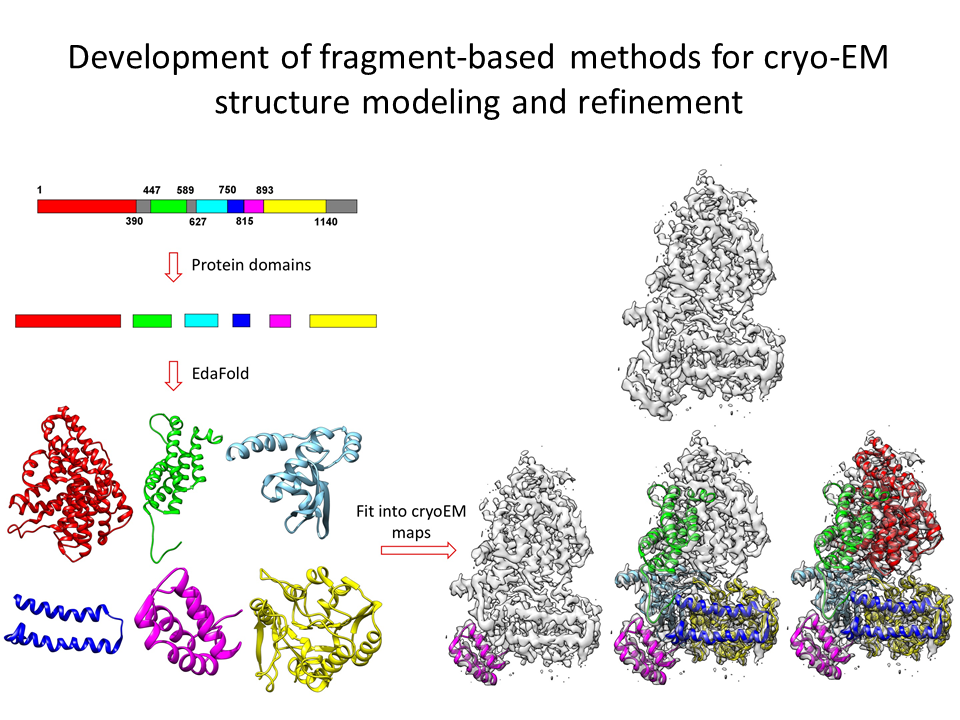
2.Development of fragment-based methods for cryo-EM structure modeling and refinement
Structural Bioinformatics Team
Team leader: Kam Zhang
We develop fragment-based modeling of structures for cryo-EM map fitting and the refinement of cryo-EM structures using fragment-assembly.
-
-
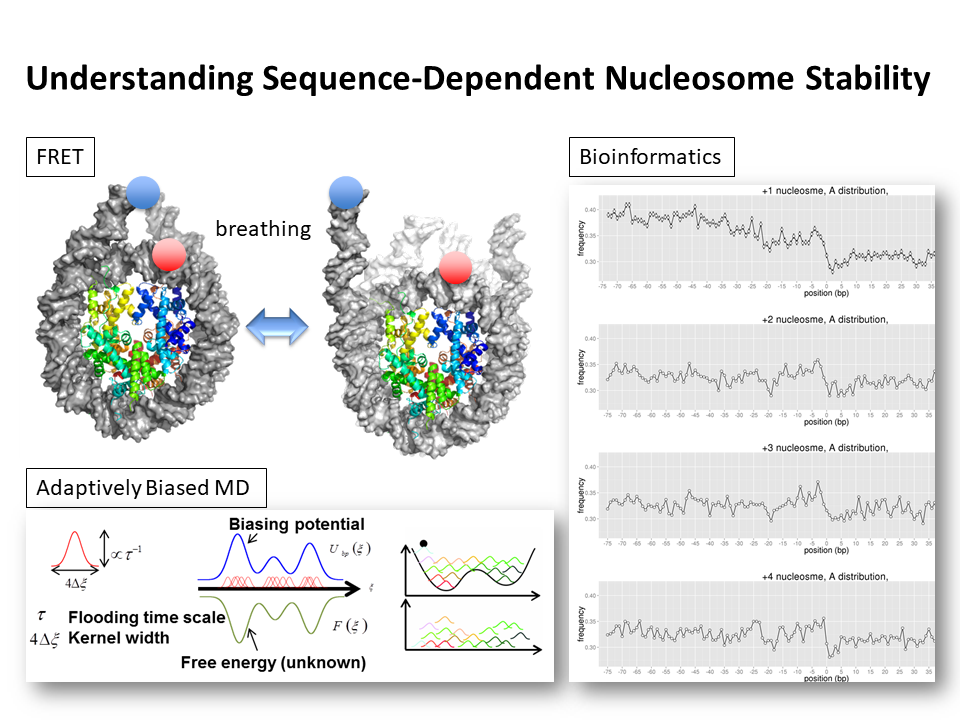
3.Understanding Sequence-Dependent Nucleosome Stability
Molecular Modeling and Simulation,
National Institutes for Quantum and Radiological Science and Technology (QST)
Team leader: Hidetoshi Kono
We try to characterize and address sequence-dependent nucleosome stability using all atom simulations, bioinformatics and FRET experiments. In particular, we focus on +1 nucleosomes and elucidate the functional role in gene regulation.
-
-
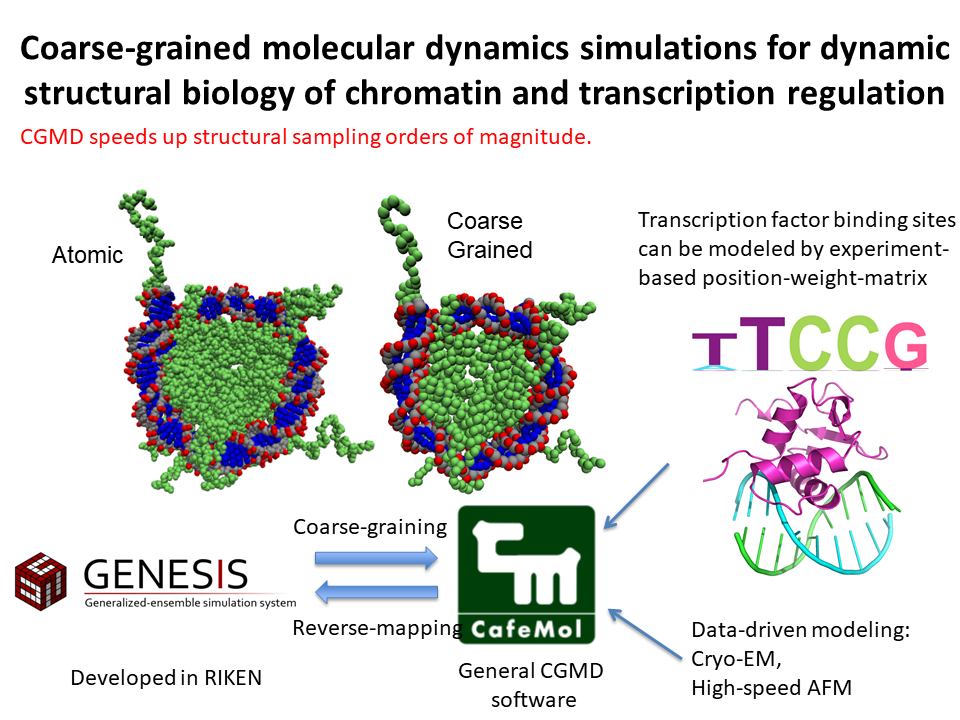
4.Coarse grained molecular dynamics simulations for dynamic structural biology of chromatin and transcription regulation
Kyoto University
Shoji Takada
We develop coarse-grained models for protein-DNA complexes and apply them to pursue dynamic aspects of nucleosome structures and their correlation with transcription activity.
-
-
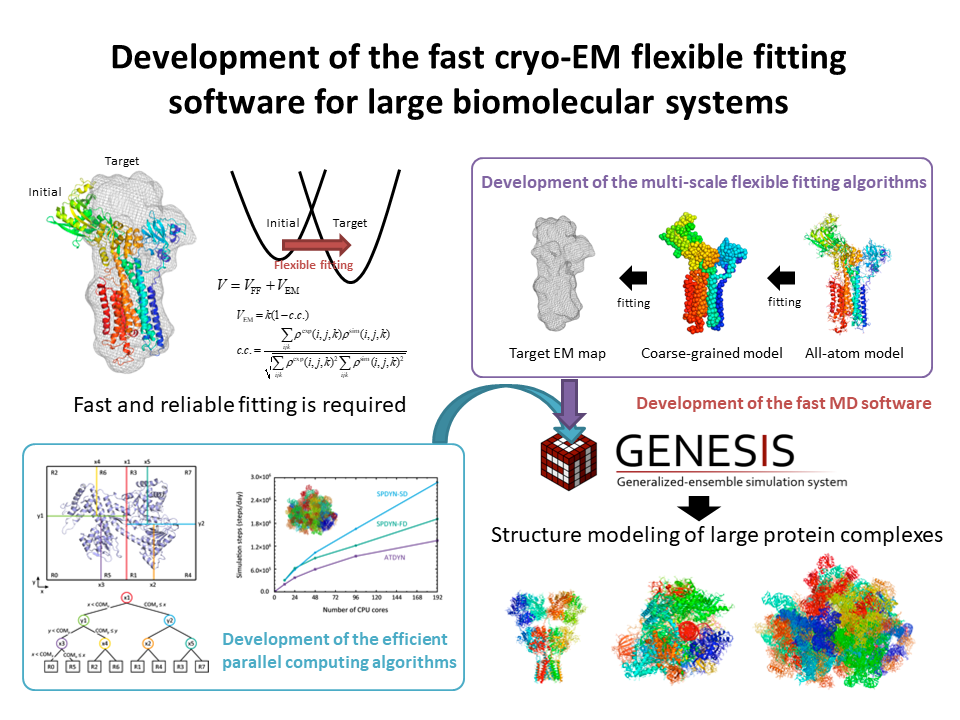
5.Development of the fast cryo-EM flexible fitting software for large biomolecular systems
Theoretical molecular science laboratory
Team leader: Yuji Sugita
We develop the new parallelization algorithm and multi-scale algorithm for the fast and reliable cryo-EM flexible fitting simulations of large biomolecular systems.
-
-
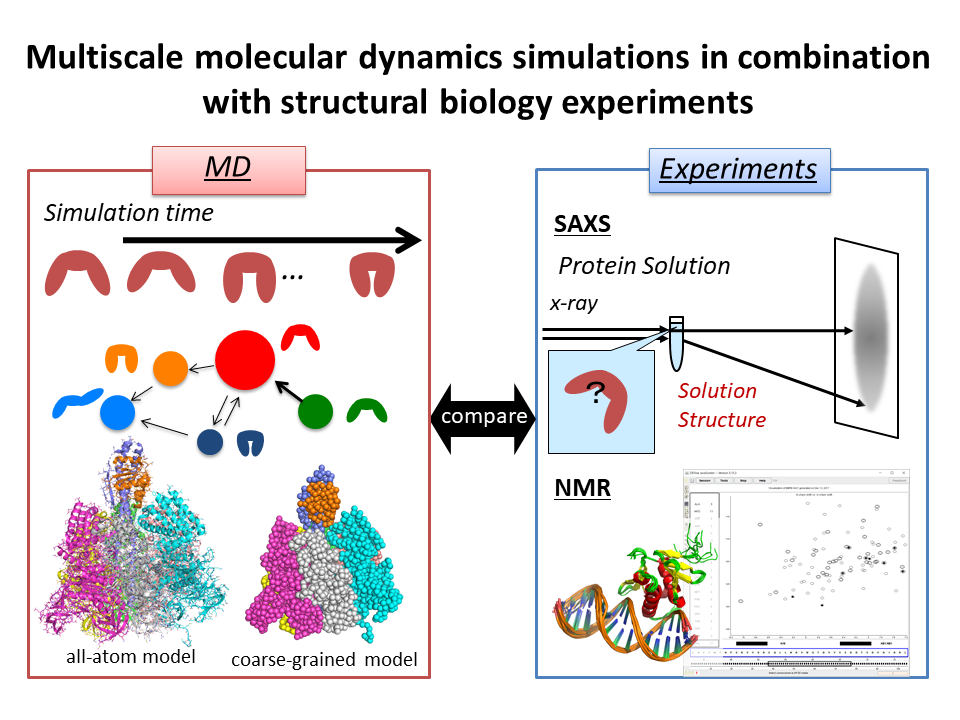
6.Multiscale molecular dynamics simulations in combination with structural biology experiments
Yokohama City University
Team leader: Mitsunori Ikeguchi
We perform multiscale molecular dynamics simulations in combination with structural biology experiments such as small-angle x-ray scattering (SAXS) and NMR.
-
-
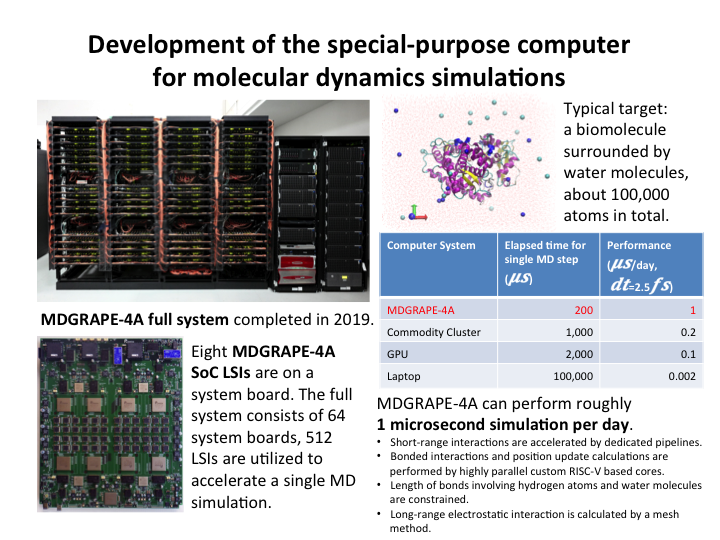
7.Multiscale molecular dynamics simulations in combination with structural biology experiments
Laboratory for computational molecular design
Team leader: Makoto Taiji
We develop the new special-purpose computer MDGRAPE-4A for all atom simulations of biomolecules and perform unbiased microseconds simulations of target proteins.

-
Sub Project 1
1.Time-resolved XFEL crystallography and vibrational spectroscopy for capturing protein dynamics at work
RIKEN SPring-8 Center, Imaging development Team
Senior Research Scientist: Minoru Kubo
We develop the time-resolved techniques of X-ray crystallography and vibrational spectroscopy, and study protein reaction dynamics at the atomic and electronic levels in real time.

-
Sub Project 1
2.Structural studies of the macromolecular complexes with chromatin
Laboratory of structural biology
Team leader: Hitoshi Kurumizaka
We reconstitute the macromolecular complexes with chromatin and determine their three dimensional structures by cryo-EM.

-
Sub Project 1
3.Development of the dynamic structural biology platform for membrane proteins
Center for Life Science Technologies, Protein Functional and Structural Biology Team
Senior Scientist: Hideki Shigematsu
We develop the new method in cryoEM single particle analysis to visualize conformational ensembles of membrane proteins in function.

-
Sub Project 1
4.Macromolecular sample production and cryo-EM analyses for dynamic structural biology
RIKEN CLST Structural Biology Group
Team leader: Mikako Shirouzu
We develop the methods for preparation of biomolecules reflecting their dynamic functional states and operate an integrated structural analysis pipeline including cryo-electron microscopy.

-
Sub Project 1
5.High-resolution single particle cryo-EM and crystallography of membrane protein complexes
Biostructural mechanism laboratory
Chief scientist: Koji Yonekura
We carry out single particle cryo-EM and crystallography of membrane protein complexes, and develop methodologies for higher-resolution cryo-EM and analyses of charge information and multi conformational states.

-
Sub Project 1
6.Cooperative conformational changes of microtubules and their functional implications
Laboratory for Cell Polarity Regulation
Team leader: Yasushi Okada
Microtubule is a dynamic cytoskeleton and playing various essential roles in living cells. To investigate their regulation, we examine the dynamic conformational changes of tubulin subunits in microtubules by using various measurement technologies.

-
Sub Project 2
1.Efficient Synthetic Route to Novel Pi-Conjugated Molecules and Development of their Function,
Hamura Lab in Kwansei Gakuin University
Team leader: Toshiyuki Hamura
The purpose of this research is development of an efficient synthetic route to various pi-conjugated systems based on the reaction integration using the reactive molecules, having potentially attractive reactivities. Three kinds of activations by heat, light, and transition metal catalyst enable the selective construction of complex structures by introducing diverse functionalities.

-
Sub Project 2
2.NMR analyses of functional dynamics of proteins / Triplet DNP application to membrane-related proteins.
Functional Structure Biology Laboratory, Graduate School of Medical Life Science, Yokohama City University
Team leader: Hideo Takahashi
We analyze functionally important but hidden dynamics of drug-target proteins by using NMR spectroscopy. We also tackle the triplet DNP application to membrane-related proteins.

-
Sub Project 2
3.Development of methods for facile preparation of pentacene-biomolecule conjugates
Graduate School of Engineering, The University of Tokyo
Team leader: Shinsuke Sando
We have taken a challenge to develop a facile method to conjugate key molecule “Pentacene” with various biomolecules

-
Sub Project 2
4.Development of the method for sensitivity-enhanced NMR spectroscopy by Triplet-DNP
Laboratory for Biomolecular Structure and Dynamics
Team leader: Takanori Kigawa
We will develop the new method for observing molecular interaction using sensitivity-enhanced NMR spectroscopy by Triplet-DNP.

-
Sub Project 2
5.Hyperpolarization of biomolecules in solution
Uesaka Spin-isospin Laboratory
Team leader: Tomohiro Uesaka
We will develop a method to hyper-polarize biomolecules using laser and microwave for sensitivity-enhanced NMR spectroscopy.

-
Sub Project 3
1.Development of new modeling algorithms combining cryo-EM data and simulation
R-CCS Computational Structural Biology Research Unit
Team leader: Florence Tama
We develop new algorithms to construct 3D models and obtain dynamical information from Cryo-EM experimental data of biological molecules

-
Sub Project 3
2.Development of fragment-based methods for cryo-EM structure modeling and refinement
Structural Bioinformatics Team
Team leader: Kam Zhang
We develop fragment-based modeling of structures for cryo-EM map fitting and the refinement of cryo-EM structures using fragment-assembly.

-
Sub Project 3
3.Understanding Sequence-Dependent Nucleosome Stability
Molecular Modeling and Simulation,
National Institutes for Quantum and Radiological Science and Technology (QST)
Team leader: Hidetoshi Kono
We try to characterize and address sequence-dependent nucleosome stability using all atom simulations, bioinformatics and FRET experiments. In particular, we focus on +1 nucleosomes and elucidate the functional role in gene regulation.

-
Sub Project 3
4.Coarse grained molecular dynamics simulations for dynamic structural biology of chromatin and transcription regulation
Kyoto University
Shoji Takada
We develop coarse-grained models for protein-DNA complexes and apply them to pursue dynamic aspects of nucleosome structures and their correlation with transcription activity.

-
Sub Project 3
5.Development of the fast cryo-EM flexible fitting software for large biomolecular systems
Theoretical molecular science laboratory
Team leader: Yuji Sugita
We develop the new parallelization algorithm and multi-scale algorithm for the fast and reliable cryo-EM flexible fitting simulations of large biomolecular systems.

-
Sub Project 3
6.Multiscale molecular dynamics simulations in combination with structural biology experiments
Yokohama City University
Team leader: Mitsunori Ikeguchi
We perform multiscale molecular dynamics simulations in combination with structural biology experiments such as small-angle x-ray scattering (SAXS) and NMR.

-
Sub Project 3
7.Development of the special-purpose computer for molecular dynamics simulations
Laboratory for computational molecular design
Team leader: Makoto Taiji
We develop the new special-purpose computer MDGRAPE-4A for all atom simulations of biomolecules and perform unbiased microseconds simulations of target proteins.

















