We aim to understand the dynamic structure-function relationships of biopolymers such as proteins by elucidating their three-dimensional structures and dynamics. We take advantage of not only conventional X-ray crystallography and solution NMR (Nuclear Magnetic Resonance) spectroscopy but also new techniques, such as XFEL (X-ray Free Electron Laser), cryo-EM (electron microscopy), and cutting-edge high-sensitivity NMR spectroscopy developed by our team. Combination of these experimental data and computational simulations advances the understanding and prediction of structural dynamics at longer time scales and embracing larger molecular systems reaching beyond the current limits. Our highly interdisciplinary researchers with background in physics, chemistry and computational science as well as structural biology tackle challenging topics in molecular biology to achieve our aim.
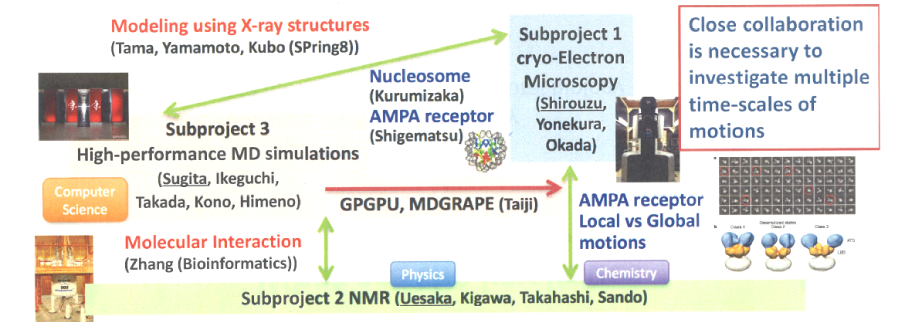
Biomolecules, such as proteins, can form various conformations or complexes by interacting with each other. It is essential to understand their dynamic structures in the living cell. Serial femtosecond crystallography or time-resolved X-ray crystallography allows us to analyze the structural changes of proteins in the range from nanosecond to second scale. In addition, the recent development of the direct-electron detector and image processing software in cryo-EM experiments enabled us to determine the three-dimensional structures of protein complexes at near-atomic resolution without crystallization. The complex structures can be determined at sub-nanometer resolution, even if various conformations are mixed in the specimen. Such analysis provides sequential information on the structural changes of proteins. In this sub-project, we combine cryo-EM experiments with molecular dynamics simulations in order to reveal the high-resolution structure of biomolecules existing in multiple conformations. Research theme in Sub-project 1
NMR spectroscopy is a versatile and non-invasive tool to determine the molecular structure and dynamics, which has shown the importance of the structural dynamics for molecular functions. NMR techniques have a huge potential in in-cell research, where biomolecules are analyzed in their cellular environments. However, the low sensitivity of NMR has been a major bottleneck in this field. DNP (Dynamic Nuclear Polarization) is the state-of-the-art technique toward high-sensitivity NMR, which enhances the signal intensities more than 104 times by increasing the nuclear spin polarization rate. In this sub-project, we use a triplet DNP (DNP using optically excited triplet states of electron spins) to achieve structural analyses at biologically-relevant temperatures and at molecular densities as close as possible to the cellular environment. In our unique group, the chemists who synthesize biomolecules with pentacene derivatives (used as a source of polarization), the physicists who develop the DNP system to enhance the nuclear spin polarization, and the biologists who analyze beautiful NMR data are working together and aiming to solve the mystery of life inside the cells.
Research theme in Sub-project 2
Molecular dynamics simulation is a useful tool to analyze structural dynamics of biomolecules. Recent advances in supercomputers enabled us to deal with a wide spectrum of biomolecular systems in terms of their size and simulation time. However, it is still difficult to predict millisecond time-scale structural changes of complex biomolecules using MD simulation alone. In this sub-project, we aim to develop new simulation techniques, which directly utilize the experimental data derived from cryo-EM, NMR and other techniques. Such a workflow prepared in tight collaboration with the experimental teams would contribute to the improvement of the resolution of predicted structures as well as understanding of slow or complicated dynamics of biomolecules. We currently focus on membrane proteins, protein-RNA complexes, nucleosome, and chromatin. Research theme in Sub-project 3